Immunotherapy has revolutionized cancer therapy, leading to the 2018 Nobel Prize in Physiology and Medicine. However, despite the dramatic response observed in several cancer types, many patients do not benefit from this treatment or relapse in a relatively short time. To improve our understanding of cancer patient response we work in several directions:
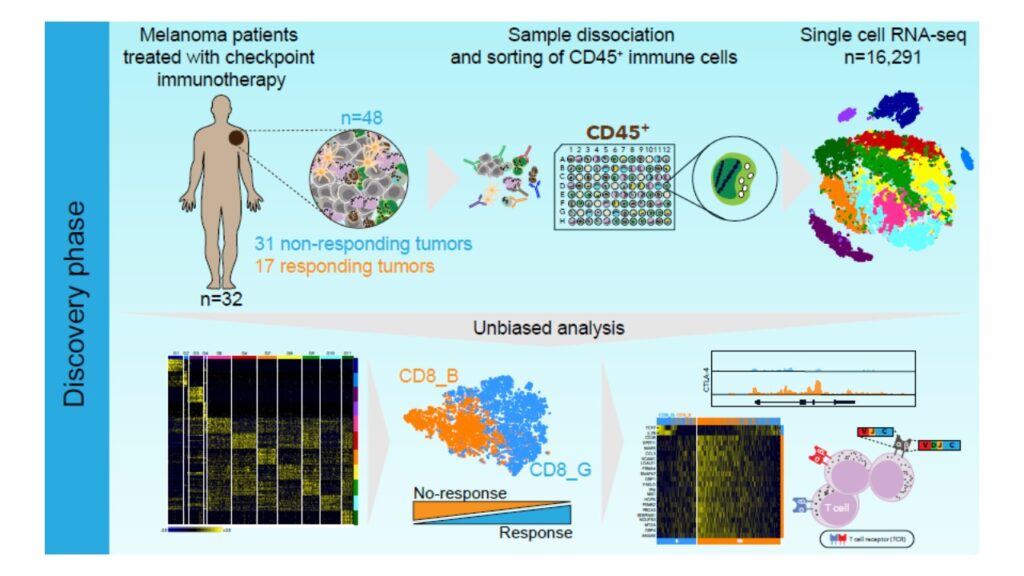
In the first, we collaborate with clinicians at Massachusetts General Hospital that collect fresh tumor samples from melanoma and lung cancer patients. These samples are processed using cutting edge technologies to generate molecular data at the single cell level. Our goal is to develop computational methods that can handle the size and complexity of these data, and address challenges related to biomarker identification, drug target prediction, and a deep characterization of the tumor’s microenvironment;
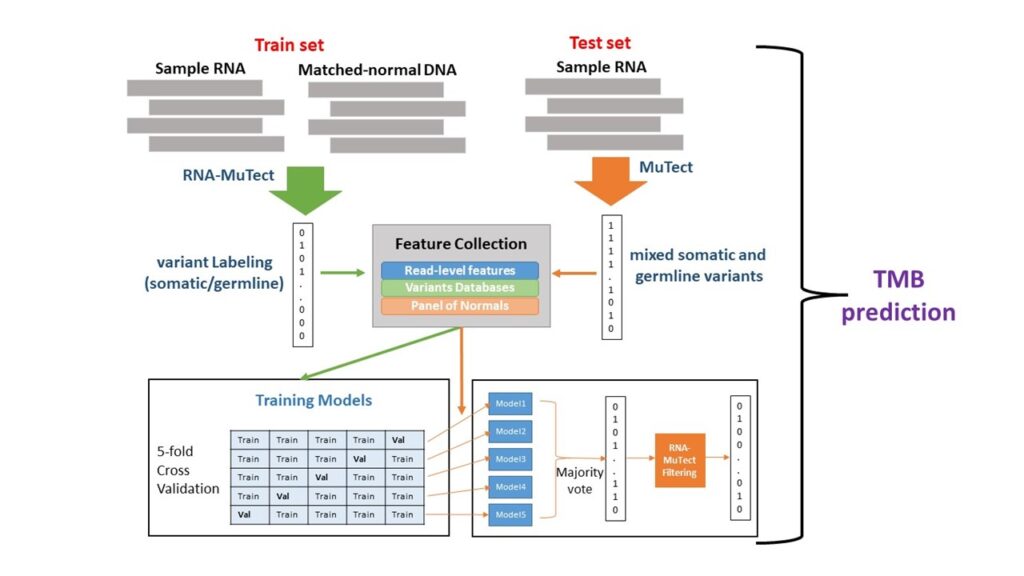
In the second, we aim to build predictors for patient response using bulk RNA sequencing data, by extracting multiple features related to the tumors' genetic and transcriptomic state. To this end, we use statistical inference and machine learning methods.
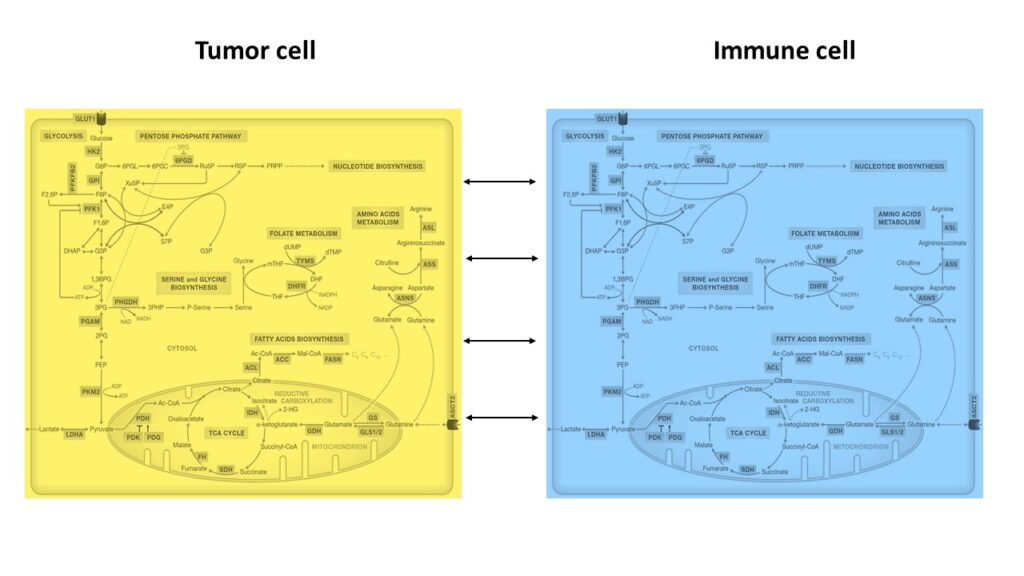
Lastly, we study the tumor-immune metabolic interactions, aiming to uncover metabolic drug targets and biomarkers that can enhance patient response. This is done via single-cell RNA-seq analysis and genome-scale metabolic modeling.